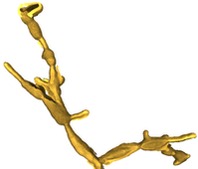
I am researching new methods to automatically detect and quantify the structure of biological specimen in stacks of microscopic images captured at incremental focal lengths. Quantifying the structure of cells and organisms is important for many biological experiments, but this process can be expensive and very time consuming when done manually. A method to automatically detect, quantify, and classify the structure of specimen in 3-D microscopic images would enable high-throughput data analysis, improved experimental efficiency and possibly lead to increased frequency of scientific discoveries.
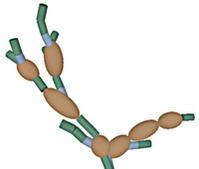
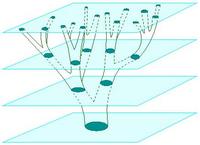
Our approach focuses on expressing the observed data as stochastically generated by an underlying model for the biological specimen and the imaging effects of the microscope. I am currently collaborating with a mycologist at the Univ. of Arizona to investigate Alternaria, a genus of fungus. We model the cellular substructure of Alternaria as 3-D geometrical primitives, such as ellipsoids and cylinders, and model its global structure as an instance of a stochastic grammar for biological growth. We accommodate the blurring effects of the shallow depth of field in the microscope by modeling its impulse response with a theoretical point spread function (PSF). This enables us to understand the image formation process and unlock structural information captured in the image blur.
Using Bayesian statistical inference, we fit both the structure and imaging models simultaneously to the microscopic images with mutual benefit; information learned from inferred structure is used to learn model parameters of the imaging system and vice versa.